2-(Isopropylamino)thieno[3,2-d]pyrimidin-4(3H)-one derivatives as selective phosphodiesterase 7 inhibitors with potent in vivo efficacy
Endo, Y., Kawai, K., Asano, T., Amano, S., Asanuma, Y., Sawada, K., Onodera, Y., Ueo, N., Takahashi, N., Sonoda, Y., Kamei, N., Irie, T.(2015) Bioorg Med Chem Lett 25: 1910-1914
- PubMed: 25866242
- DOI: https://doi.org/10.1016/j.bmcl.2015.03.031
- Primary Citation of Related Structures:
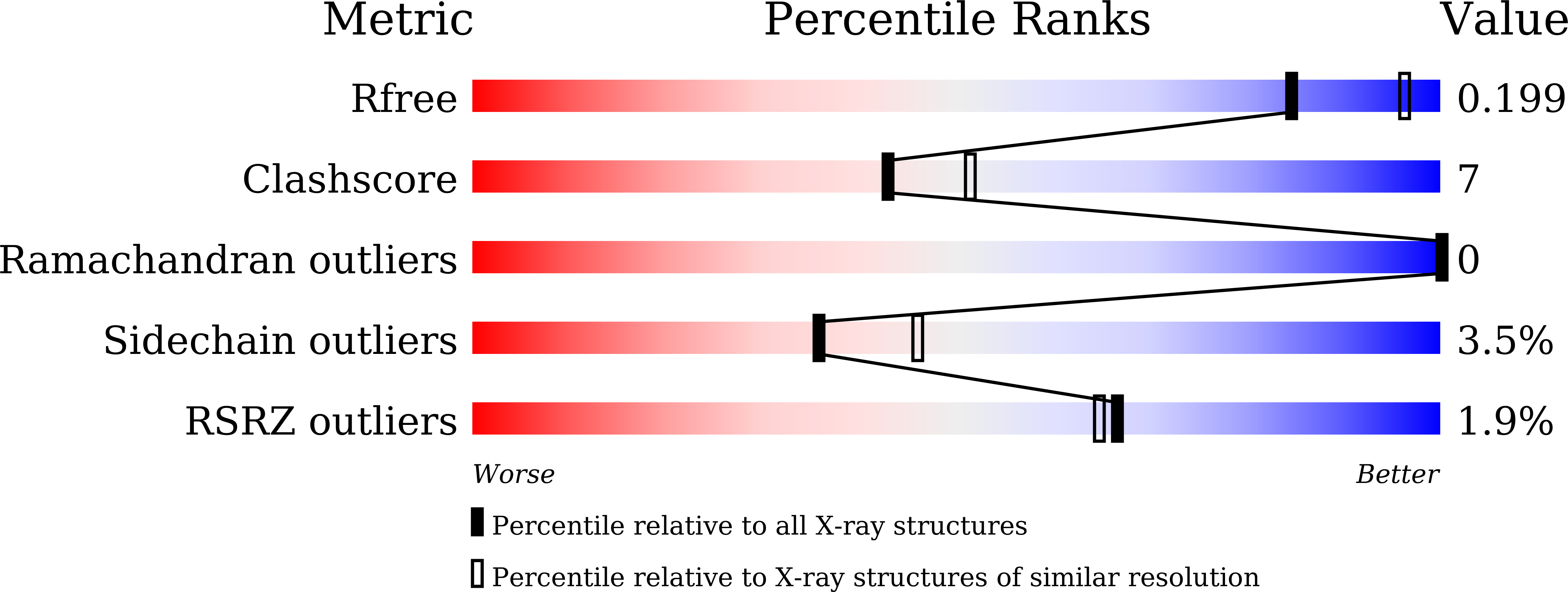
4Y2B - PubMed Abstract:
A new series of thienopyrimidinones is synthesized and evaluated as selective phosphodiesterase 7 (PDE7) inhibitors for the treatment of inflammatory diseases. The modification of the substituents on thienopyrimidinone revealed that an isopropylamino group at the 2-position was favorable for aqueous solubility. The introduction of 3-pyrrolidines at the 7-position resulted in good solubility, highly potent activity, and good PDE7 selectivity. Among the synthesized compounds, compound 46 exhibited the greatest inhibition of ear edema in a phorbol ester-induced mouse model.
Organizational Affiliation:
Drug Research Center, Kaken Pharmaceutical Co. Ltd, 14, Shinomiya Minamigawara-cho, Yamashina, Kyoto 607-8042, Japan. Electronic address: endo_yusuke@kaken.co.jp.